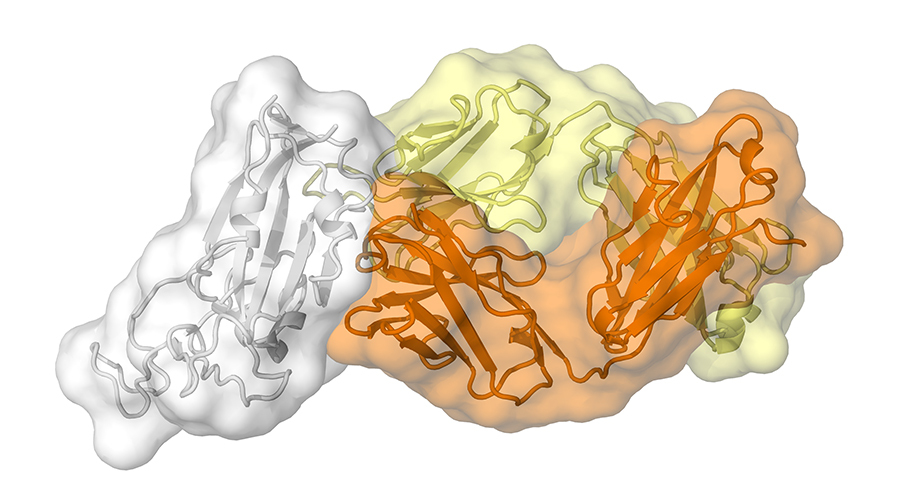
An antibody called CR3022, produced by a patient in response to the coronavirus that causes SARS, also binds to the new coronavirus that causes COVID-19. (Image courtesy of Meng Yuan and Nicholas Wu of the Wilson lab.)
Clues to COVID-19 coronavirus’s vulnerability emerge from an antibody against SARS
A Scripps Research study reveals a likely site of vulnerability on the SARS-CoV-2 virus.
April 03, 2020
LA JOLLA, CA — An antibody recovered from a survivor of the SARS epidemic in the early 2000s has revealed a potential vulnerability of the new coronavirus at the root of COVID-19, according to a study from scientists at Scripps Research.
The study, published today in Science, is the first to map a human antibody’s interaction with the new coronavirus at near-atomic-scale resolution. Although the antibody was produced in response to an infection of SARS (severe acute respiratory syndrome), which is caused by the SARS-CoV virus, it cross-reacts with the new coronavirus, SARS-CoV-2.
The structural mapping revealed a nearly identical site on both coronaviruses to which the antibody binds, suggesting a functionally important and vulnerable site for this family of coronaviruses.
“The knowledge of conserved sites like this can aid in structure-based design of vaccines and therapeutics against SARS-CoV-2, and these would also protect against other coronaviruses—including those that may emerge in the future,” says the study’s senior author Ian Wilson, DPhil, Hansen Professor of Structural Biology and Chair of the Department of Integrative Structural and Computational Biology at Scripps Research.
SARS-CoV, which causes SARS, originated in horseshoe bats, but jumped to humans in South China in 2002, eventually infecting more than 8,000 people and killing almost 800 before it was quelled by lockdowns, quarantines and other measures.
SARS-CoV-2, a closely related coronavirus that causes COVID-19, first emerged in the Chinese city of Wuhan in late 2019. Much more infectious than its viral cousin, it has led to a pandemic, causing far more cases of illness and fatalities than SARS. The development of a vaccine or even an effective treatment could significantly ameliorate the crisis.
The Wilson lab is known for its pioneering structural studies of antibodies bound to viruses including HIV and influenza. These studies have been used to inform designs of vaccines and antibody drugs, as well as other therapeutics. Along with hundreds of other labs around the world, Wilson’s team is now focused on SARS-CoV-2.
“Our ultimate goal here is to obtain structural information on antibodies and their binding sites, and use that to guide SARS-CoV-2 vaccine design, just as our lab has done with influenza and HIV,” says the study’s co-first author Nicholas Wu, PhD, a postdoctoral research associate in the Wilson lab.
The new study centers on an anti-SARS-CoV antibody called CR3022 that was originally isolated in 2006 by the pharmaceutical company Crucell Holland B.V. in the Netherlands. A report from Chinese scientists earlier this year indicated that CR3022 cross-reacts against SARS-CoV-2. Wilson’s team used their structural mapping expertise to determine how the antibody binds to SARS-CoV-2.
A key finding is that the antibody’s binding site is highly similar between the two coronaviruses—differing by just four protein building blocks called amino-acids. That high degree of similarity implies that the site has an important function that would be lost if it mutated significantly.
Yet, the site’s function remains mysterious. The Scripps Research analysis found that the antibody binding site is relatively remote from the part of the virus that grabs hold of cell-surface protein receptors in preparation for penetrating cells in our lungs. That suggests that, at least for SARS-CoV, CR3022 neutralizes the virus’s ability to infect cells in some indirect way.
Adding to the mystery is the finding that the antibody binding site on these viruses is not normally accessible to antibodies.
“We found that this region is usually hidden inside the virus, and only exposed when that part of the virus changes its structure, as it would in natural infection,” says co-first author Meng Yuan, PhD, also a research associate in the Wilson lab.
Despite the slightness of difference between the two coronaviruses, the antibody binds much less tightly to SARS-CoV-2 than it does to the SARS virus, and cannot neutralize SARS-CoV-2 in lab dish tests as it does SARS-CoV.
Still, the findings suggest that the binding site for this antibody on SARS-CoV-2 is a site of vulnerability, and that antibodies binding it more tightly would plausibly succeed in neutralizing the virus. Such neutralizing antibodies, if developed into therapies, could be used to treat COVID-19 patients and to provide temporary protection from the virus to uninfected individuals, for example healthcare workers.
The fact that this binding site is highly conserved between SARS-CoV and SARS-CoV-2 also hints that there may be antibodies, still to be discovered, that can effectively neutralize both viruses—and perhaps in the same way, can neutralize future emergent coronaviruses before they can cause pandemics.
Labs at Scripps Research and throughout the world are currently seeking antibodies, via blood donations, from people who have recovered from COVID-19 for further studies along these lines.
Authors of “A highly conserved cryptic epitope in the receptor-binding domains of SARS-CoV-2 and SARS-CoV” are Meng Yuan, Nicholas Wu, Xueyong Zhu, Chang-Chun Lee, and Ian Wilson, of Scripps Research; and Ray So, Huibin Lv, and Chris Mok of the University of Hong Kong.
Support was provided in part by the National Institutes of Health (K99 AI139445) and the Bill and Melinda Gates Foundation (OPP1170236).
For more information, contact press@scripps.edu
No comments:
Post a Comment